Predicting Growth Phenotypes by Logical Modelling
A major task of a logical model is to predict the outcome of auxotrophic experiments where the change in cell growth/division etc is measured, when one or more ORFs is/are removed(knocked out) from the Yeast genome. Traditionally these types of experiments were used only for "auxotrophic" mutants - where the ORF(s) removed resulted in no further growth of the yeast. This was the experimental regime for the rediscovery work, where the task focused on the 8 (out of 15 total) auxotrophic mutants derived from the AAA pathway.
The function of the removed gene can then be discovered by finding a chemical compound or set of compounds that restore healthy growth, since it can be inferred that the removed gene plays a role in the synthesis of the compounds, i.e. the removed gene codes for (a) reaction(s) that produce the compound(s) directly or (a) reaction(s) that produce a crucial intermediary compound.
The task of predicting growth outcomes using the metabolite graph/robot model is related to the problem of finding a path in the metabolite graph from a set of initial compounds, usually representing a growth medium and any additional compounds to test recovery of growth; to a set of compounds deemed essential for cell growth/division etc. The task is simplified by assuming that certain common compounds such as ATP, NADP etc are always present in the cell.
The reactions themselves are used to govern cell behaviour because they are a more realistic simulation of how chemical transformations occur in the cell, for a reaction to proceed all compounds in the substrates OR products need to be present in the cell before the reaction can occur:
A reaction can take place IFF:
- All of the compounds in the Subtrates list are present in the correct cell compartments
- OR all of the compounds in the Products list are present in the correct cell compartments AND the reaction is reversible
Simulation of a reaction is completed by adding the Substrates OR Products of the reaction to the correct compartments, depending on the direction i.e. the Products are added if the Substrates are present, a forwards reaction; and the Substrates are added if the reaction is run in reverse when the Products are found in the cell. A list of compounds corresponding to the contents of each cell compartment is constructed as each reaction is processed in turn. Each reaction may be run at most once. The reactions are processed in a cyclical manner until a cycle results in no new reaction executions, or all reactions have been run. At this point the cell simulation is complete and the growth outcome is predicted by examining the cell contents lists to check for the presence of the essential compounds:
IF ALL essential compounds are present, THEN the cell will continue to grow
IF ANY essential compound is missing then cell growth will be arrested (this is a prediction of an auxotrophic mutant)
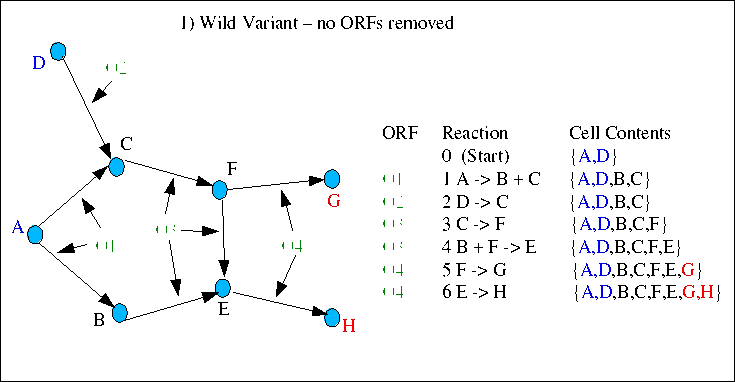
The 4 following cases illustrate hypothetical disruptions in a simple graph with 8 metabolites, 6 reactions and 4 ORFs. There are two starting compounds (A and D) and two essential compounds (end points - G and H). It includes a common situation where gene products from a single ORF catalyse more than one reaction. In these diagrams the reactions are listed in the order of execution by the simulator, and the changing cell contents list is also show. The first case is equivalent to the "wild variant" - where no ORFs have been removed, and there is a path from starts to ends.
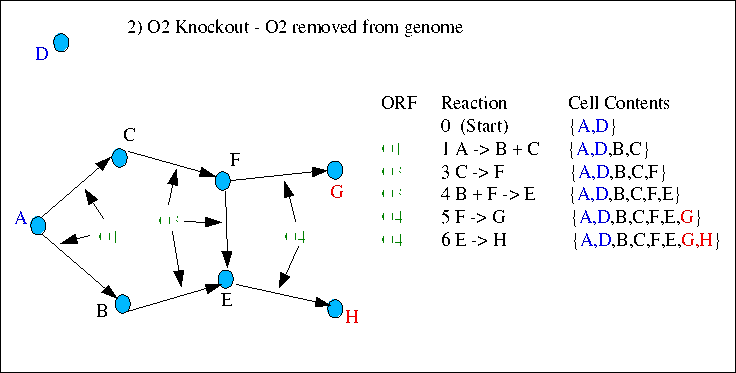
In the second case ORF O2 has been removed and reaction 2 no longer occurs. However because compound C can be synthesised in reaction 1 (from compound A), there is still a complete path from starts to ends, and in this case the prediction would be continued healthy growth.
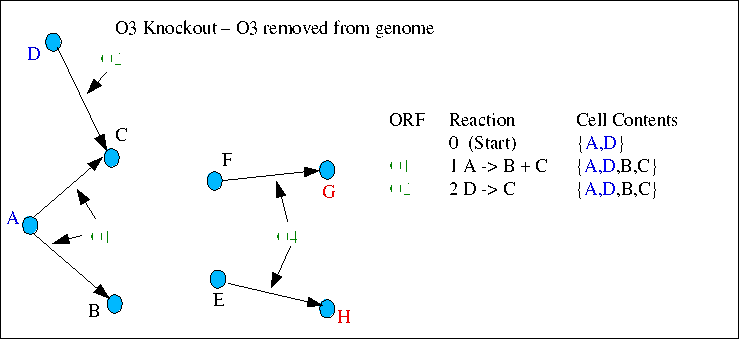
In the third case ORF O3 is removed and reactions 3 and 4 cannot take place. The cell has no mechanism for synthesis of compounds E and F, therefore neither G and H can be synthesised. Since G and H are essential compounds this case would result in an "arrested growth" prediction - i.e. this knockout mutant would be auxotrophic.
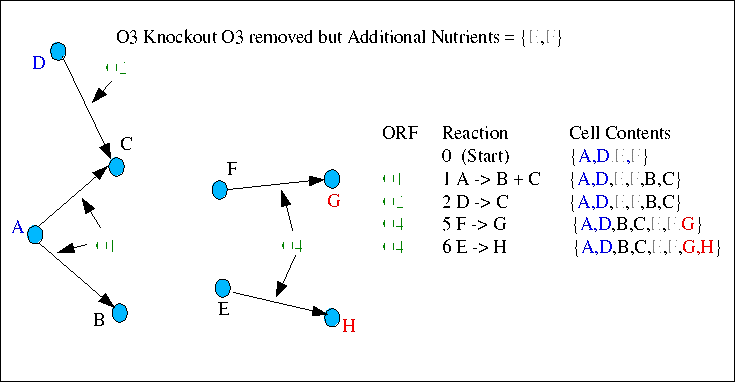
In the fourth case, ORF O3 has been removed, but the growth medium has been augmented by the addition of E and F, in an effort to identify the compounds synthesised by any reactions removed by the knocked out ORF. O3 was responsible for catalysing reactions 3 and 4, by which the cell synthesised E and F. When E and F are added to the growth medium, reactions 5 and 6 are once again functional, allowing the cell to synthesise G and H again, thereby restoring healthy growth. Since E and F are additional nutrients it can be inferred that ORF O3 plays a role in the synthesis of both compounds. Experiments and Inference of this nature can allow new reactions or chemical transformations to be discovered, as well as the genes/orfs responsible for catalysis.