sce:aber: A Logical Model of the Metabolic Network of Saccharomyces cerevisiae
This web page describes sce:aber, a logical model corresponding to the current state of knowledge about the metabolic network of of S. cerevisiae (bakers' yeast). This page will describe the construction and evolution of the model components, as well as how the model was used to generate hypotheses and experiments for execution and analysis by the Robot Scientist ADAM.
sce:aber was initially constructed and developed in 2004, as a full metabolism version of the single pathway sce:AAA model which had proved succesful as background knowledge for the proof of concept Robot Scientist work. The sce:aber model includes a more explicitly relational data representation, where the single relation used to represent the ORF, Enzyme and Reaction information comprising the AAA model is expanded to two relations: one relating reactions to the encoding ORFs/Enzymes, and one corresponding to the chemical transformations defined by the reactions. A similar but extended graph traversal algorithm to that used by sce:AAA is used to simulate the growth of wild and modified yeast strains in various environmental conditions; again focusing on the predicted outcome of auxotrophic experiments where knockout mutants are grown in a variety of growth media. Simulation has been expanded to allow the chemical composition of any defined growth medium to be used as a set of starting compounds (in conjunction with any compounds added to the growth medium to allow the experimental determination of ORF function); currently sce:aber has been used with growth media corresponding to MMD+ura+hist+leu and MMD+ura+hist+leu+meth. Determination of the predicted growth outcome remains binary (growth or no growth) but now involves testing for the prescence of a larger set of "essential compounds" i.e all the compounds deemed necessary for the continued healthy growth of S. cerevisiae. A number of "ubiquitous compounds" were also included to ensure correct simlation of the wild strain, i.e all of the essential compounds could be reached by the graph traversal algorithm.
sce:aber has been directly constructed from iFF708; a full genome yeast model designed by Foster et al [REF maybe] for Flux Balance Analysis applications; and updated by adding new information from KEGG. iFF708 splits the cell into 2 internal compartments - cytosol and mitochondrion, reflecting the prescence of both nuclear and mitochondrial DNA in eukaryote cells, and an external compartment. Transport reactions and the relevant coding ORFs are included aswell as single compartment reactions
sce:aber has an explicit representation of the reactions and ORF/enzymes as well as a PROLOG representation of the metabolic graph representing the chemical transformations. The chemical transformations are represented by two sets of PROLOG facts:
node(Location,KEGGID).
e.g. node(cytosol,'C00002').
This states that the cytosol compartment has a node
representing ATP in the graph.
reaction_edge(Location1,KEGGID1,->,Location2,KEGGID2,Reactions)
e.g. reaction_edge(cytosol,'C00002',->,cytosol,'C00008',[849,896,...]).
This states that there is a chemical transformation from ATP to ADP in
the cytosol compartment and it is found in reactions [849,896,...]
The reactions and ORFs/Enzymes are also represented by two sets of PROLOG facts:
orf_fact(Orf,Enzyme,ECFact,GeneName,GeneDesc,Reaction)
e.g. orf_fact('YBR166C','1.3.1.13,enzyme_class('1','3','1','13'),
'TYR1','prephenate dehydrogenase (nadp+),366).
This states that ORF YBR166C codes a protein belonging to enzyme
class 1.1.1.13 (also stored as a relation to allow traveral of the
Enzyme Commission class hierarchy), that the ORF is the "TYR1" gene in
Yeast, that EC class 1.3.1.13 catalyses reaction 366 which is
responsibe for prephenate dehydrogenase (nadp+) activity in the cell
Each reaction has a unique identifying number, a
direction indicator (reversible or irreversible). The Substrates and Products are
stored as lists of reactant(Location,Stoichiometry,KEGGID) where
Location is the compartment where the compound is located, Stoichiometry
is the number of atoms required by the mass balance of the reaction and KEGGID is the
6 character KEGG compound identifier:
reaction(Num,Substrates,Direction,Products)
e.g. reaction(366,[reactant(cytosol,1,'C00254'),
reactant(cytosol,1,'C00006')],->,
[reactant(cytosol,'C01179')],
reactant(cytosol,'C00011'),
reactant(cytosol,'C00005')]).
This states that Reaction 366 defines the
prephenate dehydrogenase activity coded by YBR166C, where
3-(4-Hydroxyphenyl) pyruvate is formed by removing a COOH group from
Prephenate, using NADP+/NADPH as cofactors (see below*)
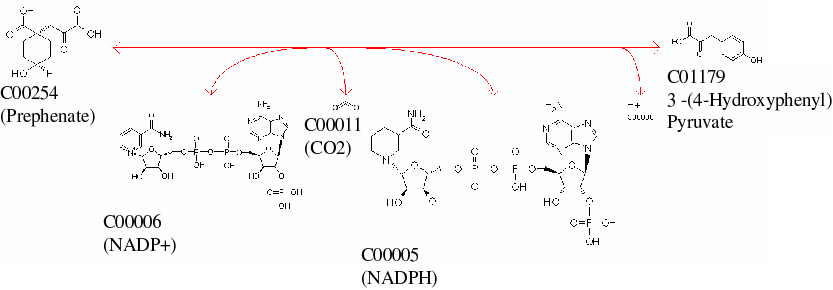
*This reaction diagram has been taken from KEGG, in the iFF708 model the H+ produced is not included and in KEGG all reactions are defined as reversible. Kegg terms this reaction as "Prephenate NADP+ Oxidoreductase (decarboxylating)"
Version History and Evolution of sce:aber
Version |
Date |
Desciption of Change |
| 1.0.0 | July 2004 | Initial Creation of Model |
| 1.1.0 | October 2006 | update of Enzyme component of supporting information for the bioinformatics method of hypothesis formation |
| 1.2.0 | October 2006 | incorporation of BLAST sequence similarity technique for bioinformatics hypthesis formation |
| 1.2.1 | June 2007 | New Growth Medium: mmd+ura+hist+leu |
| 1.2.2 | January 2008 | Reduced set of ubiquitous compounds |
Table 1 shows the evolution and version numbers of sce:aber, from the initial creation of the model (v 1.0.0) to version 1.2.2 which has a reduced set of ubiquitous compounds. Different versions have differing components, however the versioning does not necessarily refer to notions of "improvement" or "refinement", e.g Version 1.0.0 refers to the original aber model with the FASTA hypothesis generation method, the original MMD+ura+hist+leu+meth growth medium, and the original set of ubiquitous compounds, and version 1.2.0 refers to the version that used the BLAST hypothesis generation method with an updated list of enzymes from KEGG; but has no change to the growth medium or ubiquitus compounds. Version 1.2.1 was used for the comparison with iND750 and used the MMD+ura+hist+leu growth medium. Version 1.2.2 is a result of investigating whether all the original list of ubiquitous compounds was necessary for simulating the wild variety - the reduced list has only 3 compounds (ADP, O2 ad Thioredoxin) instead of the original 19. It is not possible to document or anticipate all the possible combinations of the model components. Other combinations may also be possible that have not been documented here so far, e.g. using the Blast hypothesis generation system (1.2.0) with the reduced set of ubiquitous compounds (1.2.2) and the mmd+ura+hist+leu+meth growth medium
Table 2 contains descriptions of the major components of sce:aber complete with links to KEGG so that further information regarding the ORFs, enzymes and compounds can be found. This is true for all ORFs and enzymes and 90% of compounds. Initial construction of sce:aber used the reactions from iFF708, which had a unique identification method for chemical compounds. Matches were found between this compound naming system and the KEGG naming system so that knowledge from the iFF08 surce and KEGG would be compatible; i.e. matches were found for 90% of the iFF708 compounds.| Component | Latest Version |
| Orf/Enzyme/Reaction Relations | 1.2.2 |
| Reactions | 1.2.2 |
| Complete Compound List | 1.2.2 |
| Essential Compounds | 1.2.2 |
| Original Ubiquitous Compounds | 1.2.1 |
| Reduced Ubiquitous Compounds | 1.2.2 |
| Growth Medium: mmd+ura+hist+leu+meth | 1.2.0 |
| Growth Medium: mmd+ura+hist+leu | 1.2.2 |
Table 3 is a list of the components of sce:aber, with a (downloadable) prolog file for each component. Tables 4 to 6 describe the other KEGG components used for translation (table 4), KEGG genomes used for generation of hypotheses (table 5) and the hypothesis generation methods used so far (table 6)
| Component | Latest Version |
| All ORF/Enzyme Relations | 1.2.2 |
| All Reactions | 1.2.2 |
| ORF/Enzyme Relations from iFF708 | 1.2.2 |
| Reactions from iFF708 | 1.2.2 |
| ORF/Enzyme Relationsfrom KEGG | 1.2.2 |
| Reactions from KEGG | 1.2.2 |
| Essential Compounds | 1.2.02 |
| Growth Medium: mmd+ura+hist+leu+meth | 1.2.0 |
| Growth Medium: mmd+ura+hist+leu | 1.2.2 |
| Original Ubiquitous Compounds | 1.2.1 |
| Reduced Ubiquitous Compounds | 1.2.2 |
| Model Engine | 1.2.2 |
| Component | Created (1.0.0) | Revision 1(1.1.0) |
| LIGAND Enzymes File | July 2004 | October 2006 |
| LIGAND Compounds File | July 2004 | none |
| LIGAND Reactions File | July 2004 | none |
| KEGG Web API (under development) | not used | not used |
| Component | Created (1.0.0) | Revision 1 (1.2.0) |
| KEGG Genomes (for Hypothesis generation) | July 2004 | October 2006 |
| LIGAND Reference Reactions | July 2004 | none |
| LIGAND sce Reactions | July 2004 | none |
| Component | Created (1.0.0) | Revision 1 (1.2.0) |
| Blast Hypothesis generation | none | July 2006 |
| FASTA Hypothesis Generation | July 2004 | none |